Our research: Exploring phage and Mycobacteria biology
We are a system virology lab employing a combination of interaction proteomics
and functional genomics to understand the bacterial immune response to phage
infection and the phage mechanisms responsible for immune evasion.
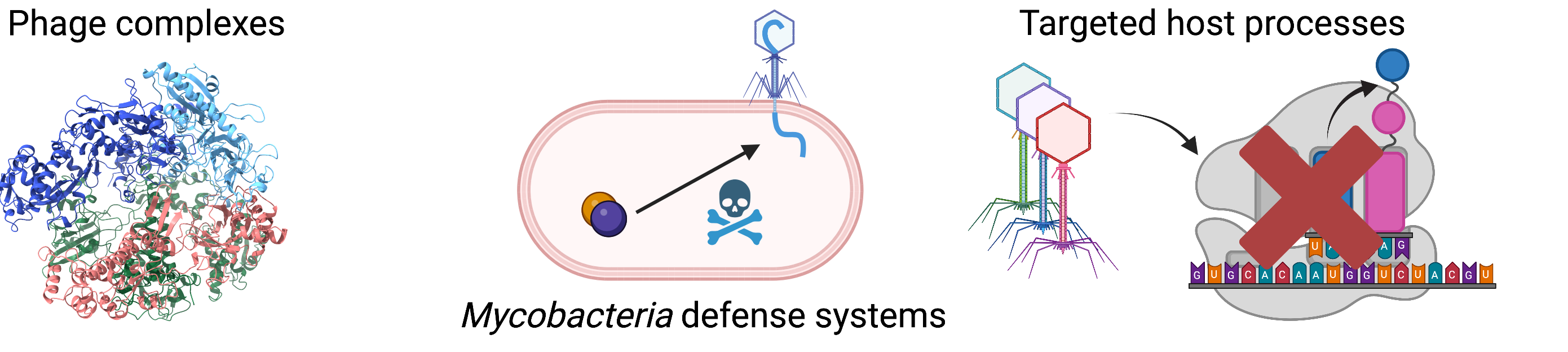
Bacterial viruses as next-generation therapeutics
Much like human cells, bacteria are susceptible to fatal viral infections. These bacterial viruses (bacteriophages, i.e phages) have gained significant attention in recent years as one of the most promising alternatives to antibiotics to tackle the emerging problem of drug-resistant bacteria.
However, just like our cells have immune systems aimed at eliminating viral
infections, bacteria possess powerful anti-phage mechanisms (like CRISPR) which greatly
reduce the efficacy of therapy using phages. Likewise, phages evolved
mechanisms to escape these systems resulting in the numerous anti-defense
systems widespread across phage families.
Our lab’s main goal is to discover these bacterial defense systems, understand their composition and
triggers as well as identifying phage mechanisms to evade them.
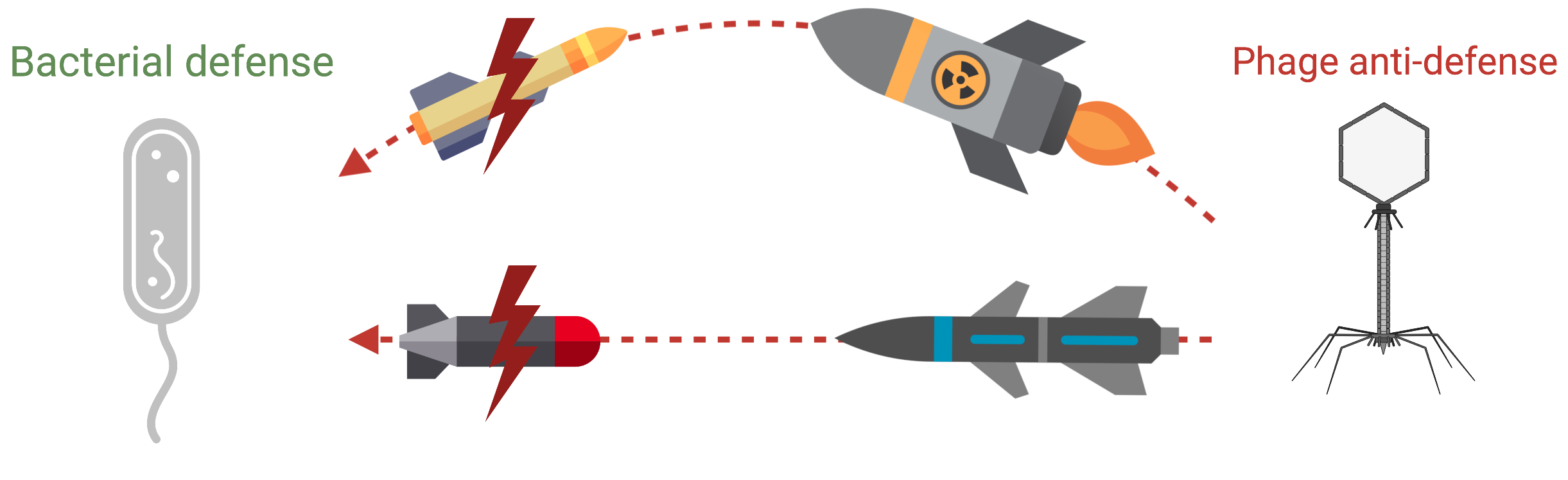
To study these systems we developed new interaction proteomics techniques for high-throughput generation of protein-protein interaction networks in phage-infected bacteria.
Co-fractionation mass-spectrometry
Most proteins are organized in macromolecular assemblies (protein
complexes), which represent key functional units regulating and catalyzing the
majority of cellular processes in health and disease. Disregulation of protein
complexes by pathogen proteins (either by direct binding or pathway-level
perturbation) is a critical determinant of infection outcome.
To identify protein complexes in a
high-throughput manner, we utilize co-fractionation mass spectrometry, a novel
interaction proteomics technique where the complexes derived from a native lysate are
size-fractionated into a large-number of fractions and each fraction is
subsequently acquired using highly quantitative mass-spectrometry (DIA-MS). The
resulting protein intensity profiles are then used as proxy for assembly state,
under the assumption that proteins sharing similar profiles were physically
associated at the separation stage.
We introduced the first machine learning model for de-novo prediction of
protein complexes from co-elution
data and continue to develop computational tools for high-throughput generation of interaction networks at scale.
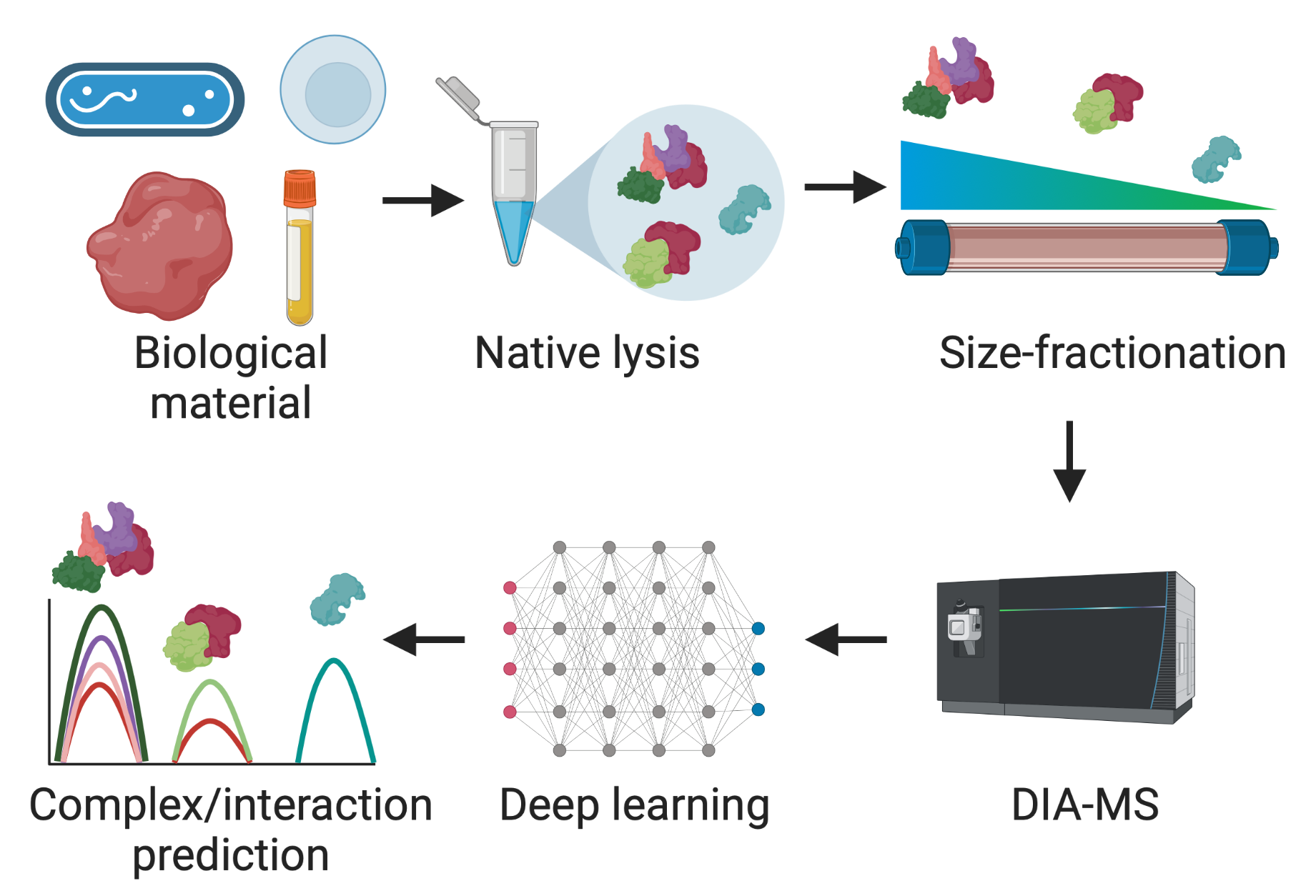 Schematic of a co-fractionation mass spectrometry experiment
Schematic of a co-fractionation mass spectrometry experiment
Interested? Reach out for an opportunity to join our team!
bfcb8cf @ 2024-06-27